Abstract
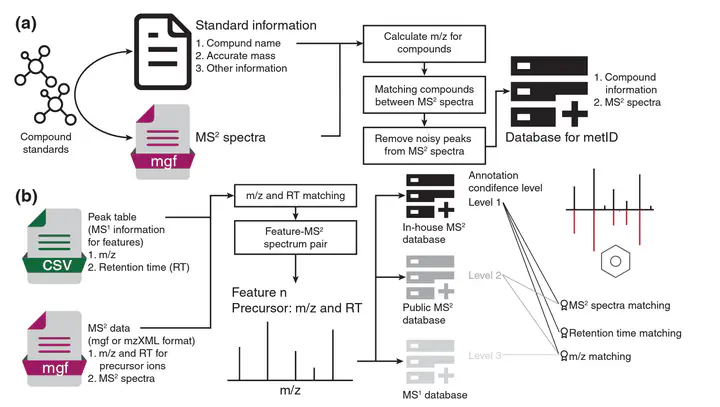
Accurate and efficient compound annotation is a long-standing challenge for LC–MS-based data (e.g. untargeted metabolomics and exposomics). Substantial efforts have been devoted to overcoming this obstacle, whereas current tools are limited by the sources of spectral information used (in-house and public databases) and are not automated and streamlined. Therefore, we developed metID, an R package that combines information from all major databases for comprehensive and streamlined compound annotation. metID is a flexible, simple and powerful tool that can be installed on all platforms, allowing the compound annotation process to be fully automatic and reproducible. A detailed tutorial and a case study are provided in Supplementary Materials.