14 Functional Module Identification
This chapter describes how to identify functional modules using the MAPA Shiny application. The Shiny app provides an intuitive interface for grouping related pathways into functional modules using optimal clustering parameters.
Prerequisites: Before identifying functional modules, ensure you have either:
- Completed pathway similarity calculation in the current session using the Pathway Similarity section as described in the previous chapter, OR
- Upload existing similarity results using the file upload option at the top of the interface
The similarity calculation results are required for functional module identification to proceed.
14.1 Overview
The Pathway Clustering section in the MAPA Shiny app provides a comprehensive workflow for identifying functional modules from pathway similarity results. It includes two main steps:
- (Optional) Find Optimal Parameters: Systematically evaluate different clustering algorithms and a series of similarity cutoffs to identify optimal parameters
- Perform Clustering: Apply the optimal parameters to generate functional modules
This two-step approach helps ensure high-quality, biologically meaningful pathway modules.
14.2 Step 1: Load Your Data
Option 1: Continue from Previous Step
If you have completed pathway similarity calculation in the current session, your data will automatically be available for clustering analysis.
Option 2: Upload Existing Results
If you have previously saved similarity results, you can upload them:
- Click “Browse” at the top of the left panel “Upload Similarity Result (.rda)” to upload your similarity results file (.rda format)
- Select your file
- Wait for validation - the app will automatically detect the input type and prepare for clustering analysis
14.3 Step 2: Find Optimal Parameters
Before performing the final clustering, it’s highly recommended to systematically evaluate different clustering parameters to find the optimal settings. The optimal parameter analysis tests three clustering algorithms:
- Binary cut: Uses the simplifyEnrichment algorithm
- Girvan Newman: Community detection based on edge betweenness
- Hierarchical: Traditional agglomerative clustering
Each method is tested across a range of similarity cutoffs to find the optimal balance between cluster cohesion and separation. You can set the evaluation parameters:
Cutoff Increment: Step size for the similarity cutoff sequence (default: 0.1)
- Smaller values (e.g., 0.05) provide more detailed analysis but take longer
- Larger values (e.g., 0.1) provide faster analysis with broader evaluation
Hierarchical Linkage (for hierarchical method): Agglomeration method options
Then run optimal parameter analysis:
- Review your cutoff increment setting
- Click the “Submit” button to start the parameter evaluation
- Monitor progress - the analysis will test multiple clustering methods and similarity cutoffs
- Wait for completion - processing typically takes 1-3 minutes depending on data complexity and cutoff increment
14.4 Step 3: Review Optimal Parameter Results
After completion, results will appear in the “Optimal Parameter Analysis” tab which includes Evaluation Plot and Best Recommended Combinations:
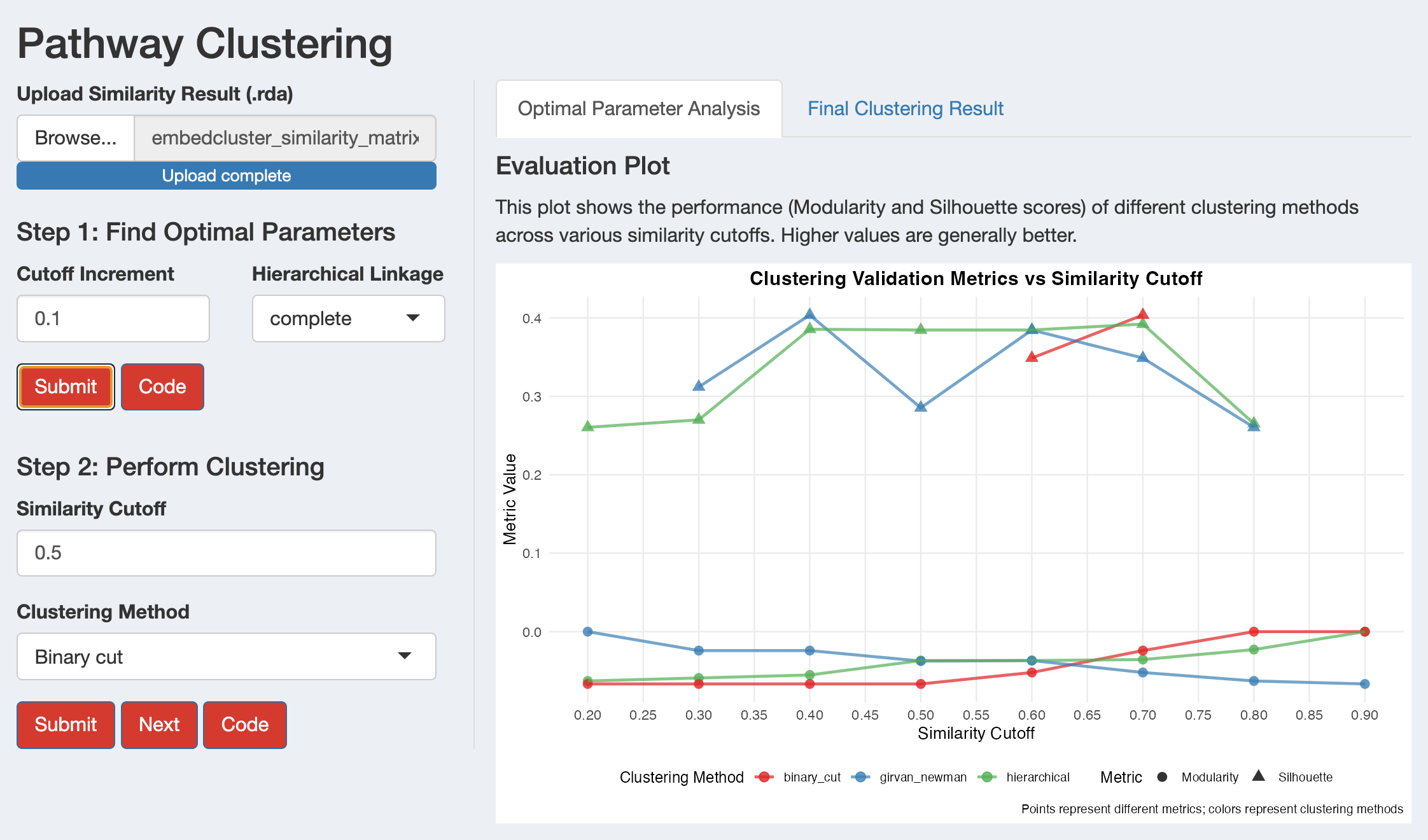
Evaluation Plot
The plot shows how clustering quality metrics change across similarity cutoffs for different clustering methods. Look for:
- Peaks in modularity: Indicate good community structure (higher = stronger community structure with clearer boundaries between clusters)
- High silhouette scores: Indicate well-separated clusters (higher = better separated clusters)
- Consistent performance: Methods that perform well across multiple cutoffs
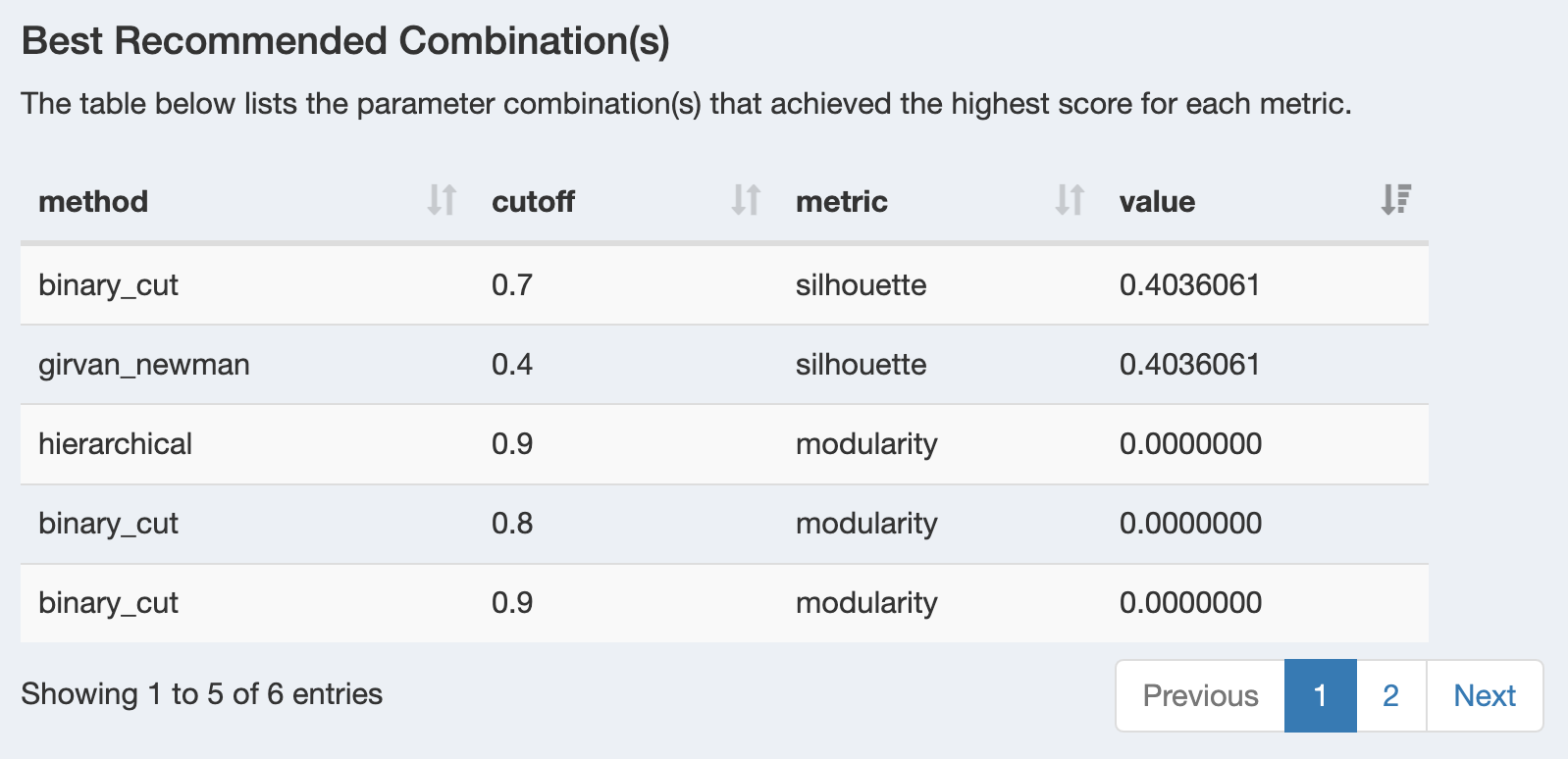
Best Recommended Combinations
The table identifies the optimal method and cutoff combinations for each metric:
- Method: Clustering algorithm (hierarchical, binary_cut, girvan_newman)
- Cutoff: Optimal similarity cutoff value
- Metric: Evaluation criterion (modularity and silhouette)
- Value: Maximum achieved score for that metric
14.5 Step 4: Run Final Clustering Parameters
Use the insights from the optimal parameter analysis to set your final clustering parameters:
Similarity Cutoff: Use the recommended cutoff from your evaluation (default: 0.5). The meaning of the cutoff depends on your clustering method:
- Girvan-Newman: Edge filter (higher cutoffs = smaller modules)
- Binary Cut & Hierarchical: Dendrogram cutting height (higher cutoffs = larger modules)
Clustering Method: Choose from the three available methods: Binary cut, Girvan Newman, and Hierarchical cluserting
Hierarchical Linkage (only for hierarchical method): Agglomeration method options
Click the “Submit” button to perform the final clustering
Monitor progress - clustering typically completes within minutes
Review results in the “Final Clustering Result” tab
14.6 Step 5: Review Final Clustering Results
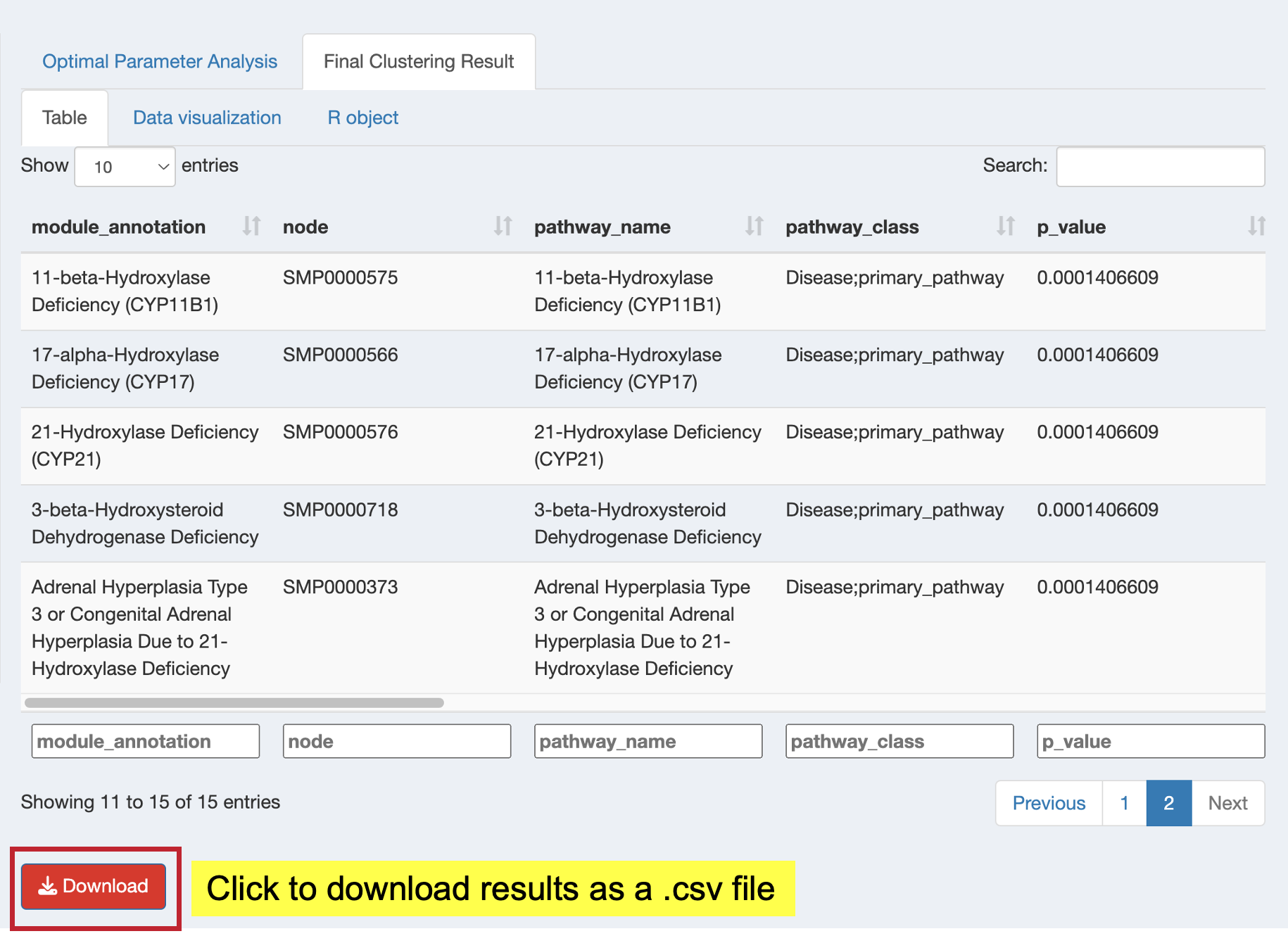
After successful clustering, examine your results in the “Final Clustering Result” tab with three sub-tabs:
Browse the functional module results and download as CSV:
- View: Complete functional module results table with key metrics
- Download: Click “Download” button to save results as CSV for further analysis
Key result columns:
- module: Functional Module identifier (e.g., “Functional_module_127”)
- module_annotation: Representative pathway name (lowest p-value for ORA, highest |NES| for GSEA)
- Description: Names of all pathways in the module (separated by
;) - module_content: All pathway/term IDs grouped in this module
- Count: Number of genes/metabolites from input list in the module
- p_adjust: Best (lowest) adjusted p-value among pathways in the module
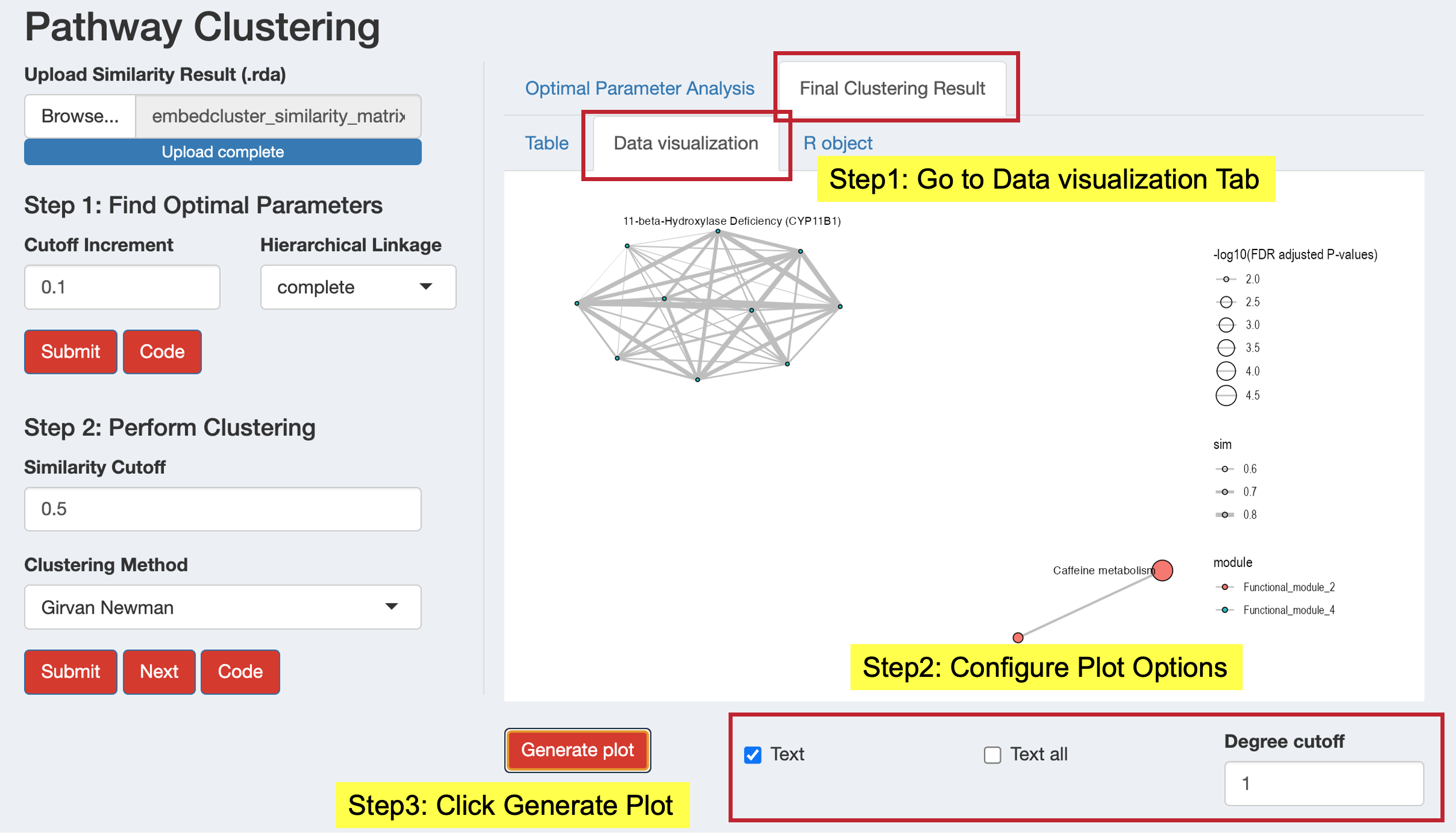
Generate similarity network plots to visualize functional modules:
- Configure plot parameters:
- Degree cutoff: Minimum pathways per module to display
- Text: Show representative module names
- Text all: Show all pathway names
- Click “Generate plot” to create the network visualization
- Examine the network to understand module relationships and structure
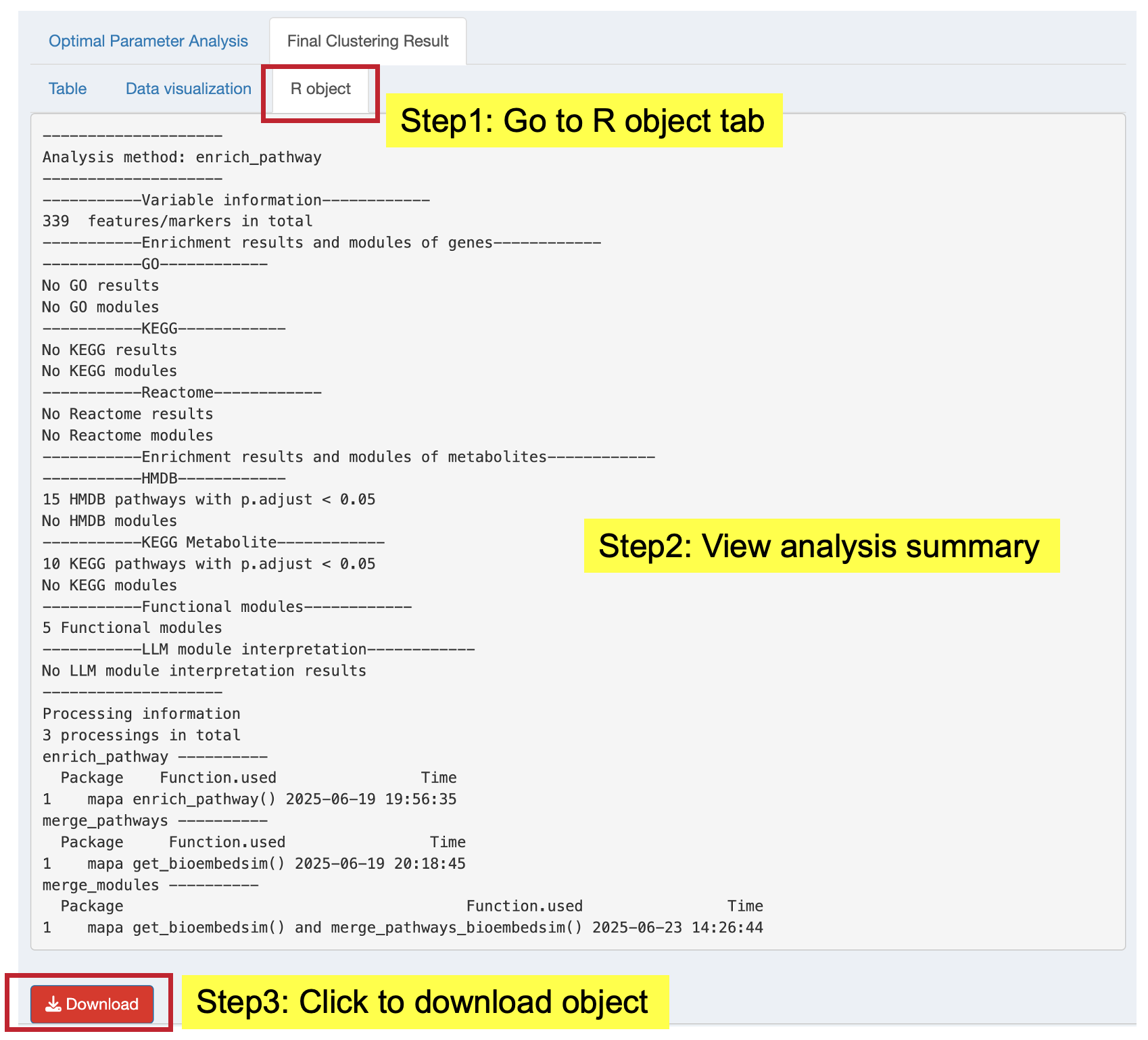
Access the complete results object for further analysis:
- View: Summary of the complete functional module analysis
- Download: Save the complete object (.rda format) for use in R
14.7 Step 6: View Analysis Code
Click the “Code” button to see the exact R code that replicates your analysis:
- Optimal Parameter Analysis Code: Shows
determine_optimal_clusters()function usage - Final Clustering Code: Shows
get_functional_modules()function usage
This code can be copied and used in your own R scripts for reproducible analysis.
14.8 Clustering Quality Assessment
Evaluate the quality of your functional modules by examining:
Module Size Distribution: Check if modules have reasonable sizes (not too many singleton modules or one dominant large module) and investigate clusters that are major outliers.
Silhouette score plot
Biological Interpretation: Most importantly, examine whether the grouped pathways represent coherent biological processes or functions. Statistical optimization alone doesn’t guarantee biological meaningfulness - modules should tell a clear biological story.
14.9 Next Steps
Once your functional module identification is complete:
- Download Results: Save both the table and R object for backup and further analysis
- Proceed to LLM Interpretation: Click the “Next” button to move to LLM Interpretation for AI-powered functional annotations of your modules
The functional modules will automatically be available for the next step in your MAPA analysis workflow, where you can add biological context and interpretations using large language models.
